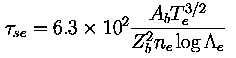Last Modified: Wednesday, 27-Oct-2021 09:26:01 BST
Current Manual for the International Multi-Tokamak Profile Database
Standard list of variables and file formats
ITPA Confinement Modelling and Database Group
 This current version of the Profile Database Manual is under development. It is the definitive source of information on Profile Database data formats etc, and over the coming months we intend to incorporate information on reading and writing MDSplus trees to bring this documentation fully up to date.
This current version of the Profile Database Manual is under development. It is the definitive source of information on Profile Database data formats etc, and over the coming months we intend to incorporate information on reading and writing MDSplus trees to bring this documentation fully up to date.
The International Multi-tokamak Profile Database is served from a computer at UKAEA Culham:
http://tokamak-profiledb.ccfe.ac.uk
The profile database stores well documented discharges that are accessible to the
fusion community through ftp, http, and MDSplus.
A discharge documentation is composed of:
- A Comments file which is a text file containing general information about the responsible
scientist who produced the data, the analysis code and miscellaneous other information.
- a 0D description file containing the global parameters of the discharge selected at some relevant time
slices corresponding to the various phases of the discharge.
- a 1D description file containing the time evolution of scalar quantities as a function of time.
- a 2D description file containing the radial profiles of quantities as a function of time and additional
information about the plasma geometry.
This document establishes what set of variables are provided for each description file together with the
definition of these variables and the standard format in which they are provided.
Once the discharge data have been submitted to the profile database, the data are stored as follow:
- via ftp or http the directory /pub/profile_data contains a list of directories corresponding to
Tokamaks from which discharges are available.
- inside the Tokamak directory is a list of directories corresponding to a particular discharge
themselves containing the comments file, 0D, 1D and 2D description file for that discharge.
- the directory /pub/profile_data
also contains a text file composed of all the 0D description files of all the discharge located on the server. It could be
used for searching discharges on the server.
This document describes, for each of the description files, the list of relevant plasma paramaters they
contain together with the file format in which they are provided.
The primary role of the profile database is to supply profile data allowing
the derivation of local transport coefficients and their comparison with
predictions from transport models. It is assumed that the transport models will
focus on thermal electron and main thermal ion transport. Therefore, the
database seeks to provide the radial profiles as a function of time of all the
relevant terms needed in the conservation equations for these two species.
Throughout this document the following definition are adopted:
The radial coordinate is the normalized toroidal flux defined as follow:
&rho=(&phi/&phia)0.5, where &phi is the total toroidal
flux enclosed by the given magnetic surface and &phia
is the total toroidal flux enclosed by the plasma separatrix (see 1D
signal # 29: PHIA). &rho goes from 0 in the center to 1 at the separatrix.
We note <.> the surface average operator defined by
< B.&nabla A > = 0 whatever A(R,Z), V(ρ) the volume
enclosed by the surface of label ρ, and S(ρ) is the corresponding
surface area.
The particle and energy conservation equations can be written:

The compressional term CMP is theory dependent and needs to be specified for
each model. For instance in the Braginskii model:
CMP=-n&alpha k T&alpha < &nabla v&alpha >
with &Gamma&alpha = n&alpha v&alpha and
where &alpha stands either for thermal electrons or for main thermal ions.
The plasma surface area is given by: S(&rho)=V'(&rho) < | &nabla &rho | >
| Quantity: | Corresponding signal number and name in this manual: |
| ϕα | 29.PHIA |
| nα |
9. NE, 38. NM1, 42. NM2, 42b. NM3, 46. NIMP |
| Tα | 1. TE, 5. TI |
| dnα/dt | 60.DNER |
| dWα/dt | 58. DWER, 59. DWIR |
| S(ρ) | 72. SURF |
| <|∇ρ|>(*) | 73. GRHO1 |
| < |∇ρ|2 >(*) | 74.GRHO2 |
| χα |
36. CHIE, 37. CHII (Note that the definition of χα does not include the <|∇ρ|2> |
(*) ρ is demensionless and the gradient is with respect to the (R,Z)
coordinates, so these quantities are in m-1 and m-2
respectively.
The comments file contains all the information users need to known about the
discharge or the way the data has been obtained but that doesn't
appear either in this manual or in the 0D, 1D or 2D files detailed in
the next chapters.
This text file should contain the following fields:
[an error occurred while processing this directive]
This information is crucial to provide users/modellers with a compact overview of the
discharges. Data providers should feel free to add in this file any information they think might be relevant to
the use of this data. Comments data should list codes and assumptions used in the data analysis. For instance EFIT for equilibrium,
PENCIL for NBI deposition profile, ONETWO for transport, "flat Zeff was assumed" or "It was asumed that Te = Ti" etc
See this example comments file
It is expected that for each discharge sent to the database one or several relevant time slices will be selected
at which time the following global data - were relevant - will be provided.
These time slices are used to summarize
the discharge and help users locate specific regimes in the database.
However, they are not necessarilly the same time slices as used in
the 2D files described below (indeed 2D Ufiles may contains hundreds
of time slices whereas for the 0D file only a few time slices will in
general be sufficient).
Additional definitions are given at the
end of this chapter for quantities used to define parameters figuring
in this standard list.
[an error occurred while processing this directive]
The following quantities were used in
the above list but not yet defined, they are not needed for the 0D
description:
WFFORM: Total fast ion energy due to NBI in joules estimated
from approximate formula. Zero if no NBI is applied.
ASDEX: From regression analysis based on 176 FREYA runs:
CFTNEL-1.3
PINJ ENBI0.75 (WTOT-WFFORM)0.5
for H beam, and C'F'T NEL-1.1
PINJ ENBI (WTOT WFFORM)0.8 for D
beam, where C and C' are estimated constants depending on the target
gas, and FT and F'T
are estimated temperature effects. Missing temperature profiles are
interpolated by regression of the available YAG temperature profiles
in the database against IP, BT, NEL, NEV, EVAP and beam gas.
D3D:

The velocities vc and vb are determined from the
critical energy and the beam energy respectively, and Pb is the
injected neutral beam power. The quantity &tause
is the slowing down-time on electrons first defined by Spitzer,
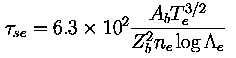
where Ab and Zb are the atomic mass and charge of the
fast ions, Te is the electron temperature in eV, ne
is the electron density in m-3, and logΛe=16
is the Coulomb logarithm. If ion drag were negligible, the term
in the brackets would be identically one. For DIII-D parameters,
however, this term varies rapidly with temperature. To give better
agreement with ONETWO results, the above formula is multiplied by
0.55.
JET:0.16*1019 PINJ/NEV for SHOT #18760, 1019(0.16
P80kV+ 0.3 P140kV + 0.02 PHe)/NEV for SHOT#
> 18760.
JFT2M: WFPER + WFPAR.
PBXM: WFPER + WFPAR.
PDX: WFPER + WFPAR.
Normal level of accuracy is ASDEX (± 15%), D3D (±
50%), JET (± 50%), JFT2M, PBXM, PDX: Co.
PFLOSS: Amount of neutral beam power in watts that is lost
from the plasma through charge exchange and unconfined orbits.
ASDEX: From fits to FREYA code results, (± 30%)
D3D: PABS exp (3.3 - IP/106)/100
(± 30%).
JET: PINJ exp (3.35 - 0.667|IP|/106 -
0.2 NEL/1019)/100 (± 50%).
JFT2M: From fits to Monte Carlo code results (± 20%).
PBXM: From a fits to the TRANSP code results (± 20%).
PDX: From a fits to the TRANSP code results (± 30%).
The required format for the 0D description follows the guidelines for the file format adopted
to the OD H mode global database and the previous suggested format.
The main difference is that the format is extendable to cope with additional variables.
The table below shows the appropriate formats and missing value codes for the three datatypes
of 0D variables.
0D Variables: types, formats, and missing values
| Type | Example 0D Variables | Format | Missing Value |
| Strings | AUXHEAT, DIVMAT, ECHLOC, TOK, WALMAT etc | A10,1X | ???????? |
| Integers | DATE, NM1A, NM1Z, UPDATE etc | I10,1X | -9999999 |
| Reals | AMIN, BT, IP, KAPPA, PNBI, Q95, TE0, ZEFF etc | 1PE10.3,1X | -9.999E-09 |
| 1X: | One blank space (ASCII code 32). |
| A10: | 10 ASCII characters. |
| I10: | Integer using up to 10 characters. |
| 1PE10.3: | Floating point number occupying at most 10
characters. Format: ±#.###E±## |
More Traditional 0D file Format
- Record length: 77 bytes
- Number of records per timeslice: unlimited (was limited to 12)
- Each record is divided into 7 fields of 11 bytes, each field is 10
characters wide and then a blank space. (i.e. I10,1X)
- The header records in the file show the names of the variable stored in each
field. These names must come from the list of allowed names given below.
- The data should be outputted for one timeslice at a time. The data
must be outputted using the appropriate format code. The format codes
to be used are given below.
- Variables missing from every timeslice do not need to be included.
- Missing values are represented as follow:
| Missing character string: | ???????? |
| Missing Integer: | -9999999 |
| Missing Real: | -9.999E-09 |
- When there is more than
one timeslice in a file the header must be repeated between each
timeslice. Each of these headers must be identical.
- The 1998 public release data always contains 78 variables.
Examples of more Traditional 0D description files:
This is an example of a 0D file in the format for the 1998 Public release. This
format is still acceptable assuming the header blocks are inserted between the
timeslices.
TOK UPDATE DATE SHOT TIME AUXHEAT PHASE
STATE PGASA PGASZ BGASA BGASZ BGASA2 BGASZ2
PIMPA PIMPZ PELLET RGEO RMAG AMIN SEPLIM
XPLIM KAPPA DELTA INDENT AREA VOL CONFIG
IGRADB WALMAT DIVMAT LIMMAT EVAP BT IP
VSURF Q95 BEPMHD BETMHD BEPDIA NEL DNELDT
ZEFF PRAD POHM ENBI PINJ BSOURCE PINJ2
BSOURCE2 COCTR PNBI ECHFREQ ECHMODE ECHLOC PECH
ICFREQ ICSCHEME ICANTEN PICRH LHFREQ LHNPAR PLH
IBWFREQ PIBW TE0 TI0 WFANI WFICRH MEFF
ISEQ WTH WTOT DWTOT PL PLTH TAUTOT
TAUTH
AUG 960215 950808 6905 2.470E+00 NBI H
STEADY 2 1 2 1 -9999999 -9999999
12 6 NONE 1.665E+00 -9.999E-09 4.700E-01 -9.999E-09
-9.999E-09 1.750E+00 1.050E-01 -9.999E-09 -9.999E-09 -9.999E-09 SN
1 ???????? ???????? ???????? ???????? -2.200E+00 1.000E+06
5.200E-01 3.570E+00 -9.999E-09 -9.999E-09 -9.999E-09 7.800E+19 0.000E+00
1.500E+00 5.000E+05 5.500E+05 6.000E+01 5.110E+06 642412 0.000E+00
-9999999 1.000E+00 5.080E+06 -9.999E-09 -9.999E-09 ???????? -9.999E-09
-9.999E-09 ???????? ???????? -9.999E-09 -9.999E-09 -9.999E-09 0.000E+00
-9.999E-09 0.000E+00 2.020E+03 2.310E+03 -9.999E-09 -9.999E-09 -9.999E-09
NONE 3.370E+05 3.690E+05 0.000E+00 -9.999E-09 -9.999E-09 -9.999E-09
-9.999E-09
TOK UPDATE DATE SHOT TIME AUXHEAT PHASE
STATE PGASA PGASZ BGASA BGASZ BGASA2 BGASZ2
PIMPA PIMPZ PELLET RGEO RMAG AMIN SEPLIM
XPLIM KAPPA DELTA INDENT AREA VOL CONFIG
IGRADB WALMAT DIVMAT LIMMAT EVAP BT IP
VSURF Q95 BEPMHD BETMHD BEPDIA NEL DNELDT
ZEFF PRAD POHM ENBI PINJ BSOURCE PINJ2
BSOURCE2 COCTR PNBI ECHFREQ ECHMODE ECHLOC PECH
ICFREQ ICSCHEME ICANTEN PICRH LHFREQ LHNPAR PLH
IBWFREQ PIBW TE0 TI0 WFANI WFICRH MEFF
ISEQ WTH WTOT DWTOT PL PLTH TAUTOT
TAUTH
AUG 960215 950808 6905 2.690E+00 NBI H
STEADY 2 1 2 1 -9999999 -9999999
12 6 NONE 1.665E+00 -9.999E-09 4.700E-01 -9.999E-09
-9.999E-09 1.750E+00 1.050E-01 -9.999E-09 -9.999E-09 -9.999E-09 SN
1 ???????? ???????? ???????? ???????? -2.200E+00 1.000E+06
3.900E-01 3.590E+00 -9.999E-09 -9.999E-09 -9.999E-09 8.000E+19 0.000E+00
1.500E+00 5.000E+05 3.570E+05 6.000E+01 7.670E+06 642412 0.000E+00
-9999999 1.000E+00 7.660E+06 -9.999E-09 -9.999E-09 ???????? -9.999E-09
-9.999E-09 ???????? ???????? -9.999E-09 -9.999E-09 -9.999E-09 0.000E+00
-9.999E-09 0.000E+00 2.680E+03 2.220E+03 -9.999E-09 -9.999E-09 -9.999E-09
NONE 4.010E+05 4.570E+05 0.000E+00 -9.999E-09 -9.999E-09 -9.999E-09
-9.999E-09
With the removal of some of the old restrictions this file can be stripped down.
The variables that are missing in all timeslices can be removed. Note that the
header blocks have been modified, but still remain fixed within the file.
TOK UPDATE DATE SHOT TIME AUXHEAT PHASE
STATE PGASA PGASZ BGASA BGASZ PIMPA PIMPZ
PELLET RGEO AMIN KAPPA DELTA CONFIG IGRADB
BT IP VSURF Q95 NEL DNELDT ZEFF
PRAD POHM ENBI PINJ BSOURCE PINJ2 COCTR
PNBI PLH PIBW TE0 TI0 ISEQ WTH
WTOT DWTOT
AUG 960215 950808 6905 2.470E+00 NBI H
STEADY 2 1 2 1 12 6
NONE 1.665E+00 4.700E-01 1.750E+00 1.050E-01 SN 1
-2.200E+00 1.000E+06 5.200E-01 3.570E+00 7.800E+19 0.000E+00 1.500E+00
5.000E+05 5.500E+05 6.000E+01 5.110E+06 642412 0.000E+00 1.000E+00
5.080E+06 0.000E+00 0.000E+00 2.020E+03 2.310E+03 NONE 3.370E+05
3.690E+05 0.000E+00
TOK UPDATE DATE SHOT TIME AUXHEAT PHASE
STATE PGASA PGASZ BGASA BGASZ PIMPA PIMPZ
PELLET RGEO AMIN KAPPA DELTA CONFIG IGRADB
BT IP VSURF Q95 NEL DNELDT ZEFF
PRAD POHM ENBI PINJ BSOURCE PINJ2 COCTR
PNBI PLH PIBW TE0 TI0 ISEQ WTH
WTOT DWTOT
AUG 960215 950808 6905 2.690E+00 NBI H
STEADY 2 1 2 1 12 6
NONE 1.665E+00 4.700E-01 1.750E+00 1.050E-01 SN 1
-2.200E+00 1.000E+06 3.900E-01 3.590E+00 8.000E+19 0.000E+00 1.500E+00
5.000E+05 3.570E+05 6.000E+01 7.670E+06 642412 0.000E+00 1.000E+00
7.660E+06 0.000E+00 0.000E+00 2.680E+03 2.220E+03 NONE 4.010E+05
4.570E+05 0.000E+00
Simpler Alternative CSV File Format
The above format for 0D files is quite complex for reading and writing the files.
A much simpler comma separated variable format is proposed with the following benefits
- simplify the format of the file
- allow creation and viewing from spreadsheets
-
| TOK | UPDATE | DATE | SHOT | TIME | … |
| JET | 20031201 | 20001006 | 53521 | 1.00E+01 | … |
| JET | 20031201 | 20001006 | 53521 | 1.10E+01 | … |
- simplify reading and writing code
The comma separated value (CSV) file format would be one line of header followed by one line per
timeslice:
TOK,UPDATE,DATE,SHOT,TIME,…
JET,20031201,20001006,53521,1.00E+01,…
JET,20031201,20001006,53521,1.10E+01,…
The only restriction would be that the strings between the commas should not exceed 10 characters. Padding with spaces is not required between commas.
Example reading and writing routines exist for Fortran, C and IDL.
[an error occurred while processing this directive]
Note that all radial profile should be given using the square root of
the normalized toroidal flux as the radial label r.
[an error occurred while processing this directive]
The file format for 2D profiles implies that a fixed radial vector in RHO
space be used for each time point given in the time vector. However,
experimental data are often measured on a grid (typically the major
radius) which does not correspond to a fixed grid when translated in
RHO space. In such cases it is not possible to project the
measurement grid onto a fixed RHO grid for each time point without
significant loss of information. To remedy this problem, an
additional level of indirection is introduced: an index is used to
label each radial measurement as a function of time, a separate 2D
file gives the RHO coordinate for each index as a function of time.
The process can be schematically represented as follows:
Standard 2D format:
RHO vector (rho1, rho2, .... rhoN)
TIME vector: (t1, t2, .... , tM)
DATA vector: (data(rho1,t1),...,data(rhoN,t1), ..., data(rhoN, tM)
Index format:
Data file:
INDEX vector (0, 1, .... N)
TIME vector: (t1, t2, .... , tM)
DATA vector: (data(0,t1),...,data(0,t1), ..., data(N, tM)
RHO file:
INDEX vector (i1, i2, .... iN)
TIME vector: (t1, t2, .... , tM)
RHO vector: (rho(i1,t1),...,rho(iN,t1), ..., rho(iN, tM)
This special format is to be used only for:
* measured quantities that would lose significant information if
projected onto a fixed RHO grid as a function of time,
* the following 2D
signals:TEXP, TEXPEB, TIXP, TIXPEB, NEXP, NEXPEB, NM1XP,
NM1XPEB, NM2XP, NM2XPEB, NM3XP, NM3XPEB, NIMPXP, NIMPXPEB,
VROTXP, VROTXPEB. (i.e. experimental measurements and their
corresponding error bars)
* The signal name for the RHO vector of the RHO file should be the name of
the signal with 'RHO' at the beginning.For instance,
experimental data points for TE would have the signal name: TEXP and
will be followed by a file with singal name: 'RHOTEXP'
Note that equilibrium data is only included in the MDSplus tree structure, and this branch closely follows the
structure which was proposed and adopted by the Pedestal Group.
EQUIL Variables
| EQUIL Signal | Units | Data Type | Brief Description | Full Description |
| EQUIL:FFPRIMPSI | T.rad | Real | FdF/dPSI
2D:- FFPRIMPSI(psi,t) | FdF/dPSI, where F=RB&phi and PSI is poloidal flux per radian
2D:- FFPRIMPSI(psi,t) |
| EQUIL:FPOLPSI | Tm | Real | FPOLPSI=(RBt)
2D:- FPOLPSI(psi,t) | Toroidal field function FPOLPSI=(RBt)
2D:- FPOLPSI(psi,t) |
| EQUIL:PPRIMEPSI | Pa.rad/Wb | Real | dP/dPSI
2D:- PPRIMEPSI(psi,t) | dP/dPSI, where P is plasma pressure and PSI is poloidal flux per radian
2D:- PPRIMEPSI(psi,t) |
| EQUIL:PRESPSI | Pa | Real | Plasma pressure,
2D:- PRESPSI(psi,t) | Plasma pressure vs. poloidal flux
2D:- PRESPSI(psi,t) |
| EQUIL:PSIBDRY | Wb/rad | Real | PSI at separatrix
1D:- PSIBDRY(t) | PSI at separatrix
1D:- PSIBDRY(t) |
| EQUIL:PSIMAG | Wb/rad | Real | PSI at magnetic axis
1D:- PSIMAG(t) | PSI at magnetic axis
1D:- PSIMAG(t) |
| EQUIL:PSIRZ | Wb/rad | Real | Poloidal flux per radian
3D:- PSIRZ(R,Z,t) | Poloidal flux per radian
3D:- PSIRZ(R,Z,t) |
| EQUIL:QPSI | - | Real | Safety factor Q
2D:- QPSI(psi,t) | Safety factor Q
2D:- QPSI(psi,t) |
| EQUIL:RBDRY | m | Real | R values on plasma boundary contour
2D:- RBDRY(index,t) | R values on plasma boundary contour
2D:- RBDRY(index,t) |
| EQUIL:RBDRY:NPTS | - | Integer | number of boundary points
1D:- NPTS(t) | number of boundary points
1D:- NPTS(t) |
| EQUIL:RCONTR | m | Real | R values on PSI contours
3D:- RCONTR(index, psi, t) | R values on PSI contours
3D:- RCONTR(index, psi, t) |
| EQUIL:RCONTR:NPTS | - | Integer | number of points on each PSI contour
2D:- NPTS(psi,t) | number of points on each PSI contour
2D:- NPTS(psi,t) |
| EQUIL:RMAXIS | m | Real | R at magnetic axis
1D:- RMAXIS(t) | R at magnetic axis
1D:- RMAXIS(t) |
| EQUIL:RXPT | m | Real | R at X-point(s)
2D:- RXPT(X-point-index,t) | R at X-point(s)
2D:- RXPT(X-point-index,t) |
| EQUIL:RXPT:NPTS | - | Integer | number of X-points
1D:- NPTS(t) | number of X-points
1D:- NPTS(t) |
| EQUIL:ZBDRY | m | Real | Z values on plasma boundary contour
2D:- ZBDRY(index,t) | Z values on plasma boundary contour
2D:- ZBDRY(index,t) |
| EQUIL:ZBDRY:NPTS | - | Integer | number of boundary points
1D:- NPTS(t) | number of boundary points
1D:- NPTS(t) |
| EQUIL:ZCONTR | m | Real | Z values on PSI contours
3D:- ZCONTR(index, psi, t) | Z values on PSI contours
3D:- ZCONTR(index, psi, t) |
| EQUIL:ZCONTR:NPTS | - | Integer | number of points on each contour
2D:- NPTS(psi,t) | number of points on each contour
2D:- NPTS(psi,t) |
| EQUIL:ZMAXIS | m | Real | Z at magnetic axis
1D:- ZMAXIS(t) | Z at magnetic axis
1D:- ZMAXIS(t) |
| EQUIL:ZXPT | m | Real | Z at X-point(s)
2D:- RXPT(xpoint_index,t) | Z at X-point(s)
2D:- RXPT(xpoint_index,t) |
| EQUIL:ZXPT:NPTS | - | Integer | number of X-points
1D:- NPTS(t) | number of X-points
1D:- NPTS(t) |
The 1D and 2D description files are themselves composed of a list of
files, one for each of the variables listed above.
- The format for the variable files is ASCII UFILE as developed at PPPL by D. McCune.
- 2D UFILEs should be provided as a function of the square root of the normalized toroidal flux (from 0 at the center to 1 at the edge).
The first independent variable should be the radial vector followed by the time vector:
RHO ;-INDEPENDENT VARIABLE LABEL: X-
TIME SECONDS ;-INDEPENDENT VARIABLE LABEL: Y-
TE ;-DEPENDENT VARIABLE LABEL-
For index file (see VI.d) the vectors are:
INDEX ;-INDEPENDENT VARIABLE LABEL: X-
TIME SECONDS ;-INDEPENDENT VARIABLE LABEL: Y-
TEXP ;-DEPENDENT VARIABLE LABEL-
and a separate set:
INDEX ;-INDEPENDENT VARIABLE LABEL: X-
TIME SECONDS ;-INDEPENDENT VARIABLE LABEL: Y-
RHOTEXP ;-DEPENDENT VARIABLE LABEL-
The radial vector RHO must be sorted in increasing order of the
normalized toroidal flux. In order words, it should increase
monotonically from the minimum value (closest to the axis) to its
maximum value (closest to the edge or even beyond the separatrix if
available).
Of course the data vector (containing the dependent variable) should
be consistent with the order in which the radial vector is given.
Similarly, the INDEX vector should contain integers in increasing
order starting from 0.
The field '-DEPENDENT VARIABLE LABEL-' should contain only the name
of the variable as it appears in this manual.
For instance:
QNBII ;-DEPENDENT VARIABLE LABEL-
Software information for reading and writing UFILEs and various
software for extracting and retrieving the data are provided,
whenever available, in: /pub/profile_data/software.
-The various files are assembled together in two single files - one
regrouping all 1D Ufiles the other regrouping all 2D Ufiles. UFILEs
are separated by two lines of at list 10 '*' (ASCII code 42) as
follow:
--- UFILE 1 -----
************************************************
************************************************
--- UFILE 2 -----
************************************************
************************************************
--- UFILE 3 -----
The file containing the separatrix information should be grouped with the 2D UFILEs.
-The comments file for the discharge (plain ASCII text) is called:
tok_#####_com.dat
The file containing the 0D data is called:
tok_#####_0d.dat
The file grouping all 1D files is called:
tok_#####_1d.dat
The file grouping all 2D files is called:
tok_#####_2d.dat
The optional files containing all measured experimental data (see section on Index method):
tok_#####_2dexp.dat
where tok is the name of the Tokamak and ##### is the pulse number.
-On the file server, the data
corresponding to the tokamak tok , shot number #####
appears in the directory: /pub/profile_data/tok/##### which
contains:
tok_#####_com.dat
tok_#####_0d.dat
tok_#####_1d.dat
tok_#####_2d.dat
-All 0D data form all discharges
figuring on the server are also grouped together in a file called
tokamak_0d.dat figuring in the directory: /pub/profile_data.
Current Manual for the ITPA International Multi-Tokamak Profile Database
Access to Restricted ITPA Profile Databases
| Database |
Working DB |
ITB DB |
| Access rules |
Available to members or approved active participants in the ITPA Confinement Database and Modelling Group |
Available to members or approved active participants in the ITPA Transport and Internal Transport Barrier Physics Group |
| Assuming eligibility, do you need to acquire... |
|
|
Data-Providers only!
Do you require... |
|
|
PR98 Data now Accessible via MDSplus
Profile Database Access via MDSplus
Profile database data is accessible from `tokamak-profiledb.ccfe.ac.uk' via MDSplus, thanks to Tom
Fredian, Josh Stillerman and Martin Greenwald at MIT who have helped with setting up the server and in
converting the data.
We'd appreciate MDSplus users testing this new way to access Profile Database data.
Please Report any Problems, so we can fix them!
Documentation on MDSplus for Profile Database:
If you are new to MDSplus, then Tom Fredian's talk is probably the best place to start.
Example routines to Read/Write MDSplus trees:
- Simple IDL Example reading a few database items into an IDL program
- IDL procedure to read all profile database data from a given discharge into IDL structures (originally from Martin Greenwald)
- Fortran Example reading a few database items into a Fortran program, with the makefile to compile with the Absoft f77 compiler (originally from David Mikkelsen)
- MATLAB Example reading a few database items into a MATLAB program (from Frederic Imbeaux)
- IDL example to write an MDSplus tree on the tokamak-profiledb.ccfe.ac.uk MDSplus server (Malcolm Walters)
- TCL script to generate profile database model tree files (Josh Stillerman):
NB you must first install MDSplus on your machine before running these!
Please also feel free to contribute any additional examples which may be of general use!
MDSplus Server and Tree Names
If you want to get started, then the following information is crucial.
1 where tok= `aug', `cmod', `d3d', `ftu', `jet', `jt60u', `rtp', 't10', 'tftr', 'ts', 'txtr'
2 permission must be granted from the appropriate ITPA group.
3 only bona-fide data-providers will be granted write access int the database transit areas
Registration for MDSplus Access to Restricted Databases
MDSplus access to WDB and ITB databases is ONLY possible for registered IP addresses/usernames. PR98 is NOT subject to this restriction. To access the restricted databases (WDB, ITB) via MDSplus, you must register with us the following information:
- the username, hostname, and IP address for the machine account from which you want MDSplus access to the profile database
- the particular database trees that you want to access: ie wdb or itb
To register simply email your registration details.
How to Contribute Data to the ITPA Profile databases
How to Contribute Data to the ITPA Profile Databases?
The International Multi-tokamak Confinement Profile Database
provides a convenient archive for any tokamak data which are suitable for
transport analysis: ie ideally this should include measured profiles of
temperatures, densities, and heat/particle sources, and estimates of
the errors in these quantities.A discharge can be submitted to the profile database either (i) by
filling the appropriate MDSplus tree, or (ii) by providing four
associated ASCII files: comments file, 0D file, 1D file, and 2D file.
The comments provide a brief description of the discharge and generally
useful auxiliary information; the 0D data provide global quantities at
one or more time points in each discharge; the 1D data give time traces
of a number of global quantities; and the 2D data gives radial profiles
of local plasma quantities at one or more time points. A full description of the profile database requirements is given in
the database manual .
If you have data which would be of interest and would like to
contribute to the profile database, please assemble your data in files as
described in the profile database manual and follow the submission
procedure which is outlined below.
The new way of submitting data is very much data-provider driven!
Only registered people can contribute new data to the database. If you
wish to submit data, please register simply by emailing
your own computer account details.
Any new data must be checked before being added to the live database.
To make sure this occurs the submission procedure has four stages.
- Data-provider places of data on the server in their
private transit area, either via FTP or via MDSplus
- Data-provider runs simple checks on the uploaded data
- Data-provider reads check logs, and flags satisfactory
discharges for upload from the transit area to the live database.
- System automatically uploads discharges which have been checked
and flagged by the data-provider from the private transit
area into the live database.
1. Placing Your Data on the Server Incoming Transit Area
Using MDSplus
Data can be written directly to the
database server by using MDSplus. A
description of the tree structure is here,
and other MDSplus information is here.
Addition of data is done using a series of calls similar to the
following
IDL code. The treename to use will be given to you on registration.
pro mdswrite
;
; IDL example to write new stripped down MDSplus tree at tokamak-profiledb.ccfe.ac.uk
; Example shows filling each node type: comments, zerod, oned, and twod
;
; Colin Roach + Malcolm Walters March 2004
;
; First connect to the server
;
mdsconnect,'tokamak-profiledb.ccfe.ac.uk'
;
; pick tree and shot number
; eg this example is to submit a d3d discharge to the working profile database
; ( to submit aug discharge to the itb database set tree='i_itb_aug' )
; nb incoming treename has 'i_' as prefix before the profile database treename
;
;###############################################################################
; strangely MDSplus needs shot number >= 100 to avoid padding with zeros
;###############################################################################
;
tree='i_d3d' & shot=100
;
; check if the tree already exists in the transit area
;###############################################################################
; nb for bizarre reasons we need to call mdsopen even if the tree does not exist!
;###############################################################################
;
mdsopen,tree,shot,status=status
;
; if the tree is already there, go straight to adding data
if (status) then goto,adddata
;
; otherwise copy the model tree
;
mdstcl, 'set tree i_d3d',status=status
print,"mdstcl, 'set tree '+tree",status
mdstcl, 'create pulse '+strtrim(shot,2),status=status
print,"mdstcl, 'create pulse'",status
;###############################################################################
; nb for bizarre reasons we need to reconnect (otherwise get mdsvalue errors)
;###############################################################################
;
mdsconnect,'tokamak-profiledb.ccfe.ac.uk'
; Open the new tree
;
mdsopen,tree,shot,status=status
print,"mdsopen",status
adddata:
print,'Add data'
; Insert the data
; First some comments
MdsPut, '\top.COMMENTS:DESCRIPTION', "$", 'Test of MDSplus writing'
print,"mdsput setting comment description = ",mdsvalue('\top.comments:description')
stop
; Then some 0D data
MdsPut, '\top.ZEROD:TOK', "$", 'D3D'
MdsPut, '\top.ZEROD:TIME', "$", 100.0
MdsPut, '\top.ZEROD:IP', "$", 50.0
print,"mdsput setting various 0D nodes "
stop
; Now some 1D data
ip_time=dindgen(1000) & ip=1.0d6*(1.0d0-exp(-(ip_time)/100))
plot,ip_time,ip,xtitle='time',title='current',charsize=2
MdsPut, '\top.ONED:IP','build_signal(build_with_units($,$),*,build_with_units($,$))',IP,'Amps',IP_TIME,'Seconds'
print,"mdsput setting various 1D IP node"
stop
; some 2D data
;
nt=10 & nx=50
x=dindgen(nx)/double(nx-1) & t=dindgen(nt)
ne0=1.0d19*(1.0d0-x^2)
dene=dindgen(nx,nt)
for i=0,nt-1 do dene(*,i)=ne0*(t(nt-1)^2-0.5d0*(t(i)-t(nt-1))^2)
surface,dene,x,t,xtitle='x',ytitle='time',title='Density',charsize=2
MdsPut, '\top.TWOD:NE', 'build_signal(build_with_units($,$),*,build_with_units($,$),build_with_units($,$))',dene,'m-3',x,'rho',t,'Seconds'
print,"mdsput setting the 2D NE signal"
EQU:
; add PSIRZ to the equilibrium branch
;
nr=32 & nz=32 & nt=3
r=dindgen(nr)/double(nr-1) & z=dindgen(nz)/double(nz-1)-0.5d0 & t=dindgen(nt)
psirz=dindgen(nr,nz,nt)
for it=0,nt-1 do begin
for iz=0,nz-1 do begin
psirz(*,iz,it)=t[it]*exp(-((r-0.5d0)^2+z[iz]^2))
endfor
endfor
!p.multi=[0,2,2]
surface,psirz[*,*,0],r,z,xtitle='R',ytitle='Z',title='PSIRZ',charsize=2
surface,psirz[*,*,1],r,z,xtitle='R',ytitle='Z',title='PSIRZ',charsize=2
surface,psirz[*,*,2],r,z,xtitle='R',ytitle='Z',title='PSIRZ',charsize=2
;
MdsPut, '\top.EQUIL:PSIRZ', 'build_signal(build_with_units($,$),*,build_with_units($,$),build_with_units($,$),build_with_units($,$))',psirz,'Wb/rad',r,'R (m)',z,'Z (m)',t,'t (s)'
print,"mdsput setting signal EQUIL:PSIRZ"
; Close this pulse
close:
mdsclose
;Since we aren't doing any more pulses disconnect.
mdsdisconnect
end
Using FTP
If you wish to submit data in ASCII files
then you can transfer them by FTP onto the database machine.
Each discharge in the database should
have four associated files: comments file, 0D file, 1D file, and 2D
file.
The comments file ("*_com.dat") gives a brief description of the
discharge and generally useful auxiliary information; the 0D file
("*_0D.dat") gives 78
global quantities at one or more time points in each discharge; the 1D
file ("*_1D.dat") gives time traces of a number of global quantities;
and
the 2D file ("*_2D.dat") gives radial profiles of local plasma
quantities at one or more time points. The 1D and 2D files are stored
in the familiar ufile
format, and a full description of the profile database format and
requirements is given in the database
manual .
To create the ASCII files for
uploading;
- Check this software link for any
software that might already exist to extract the data on your system.
- Fill in the 0D data in the format described in the 0d File Format section, and name the
file: tok_#####_0d.dat.
- Find in your local DataBase the available time traces
corresponding to the
signal list in the section on Minimal
List of 1D variables.
Translate each signal into a 1D UFILEs where the name figuring in the
manual is
used in the UFILE variable description field.Combine all 1D UFILEs into
one file
called tok_#####_1d.dat by separating each UFILE by two lines
of at least
10 '*'
- Find in your local DataBase the available radial profiles as a
function of
time corresponding to the required 2D signal
list.
Translate each signal into a 2D UFILEs where the name figuring in the
manual is
used in the UFILE variable description field, the radial coordinate is
the
square root of the normalized toroidal flux (radial vector before time
vector).
Combine all 2D UFILEs and the separatrix file into one file called tok_#####_2d.dat
by separating each UFILE by two lines of
at least 10 '*'
- Create the comments data file as detailed in section Comments Data and name the file: tok_#####_com.dat.
After creating all the data it can then
be copied onto the server.
Each tokamak has its own FTP area, each of these has an identical
layout.
An example session is shown below. Some of the server responses have
been removed for clarity.
- First connect to the server using the username and password given to you.
>ftp tokamak-profiledb.ccfe.ac.uk
Connected to tokamak-profiledb.ccfe.ac.uk
(194.81.223.118).
Name (tokamak-profiledb.ccfe.ac.uk:): jet
Password:
230-aftpd:Connected to 194.81.223.118. Logging in...
(NB depending on your own firewall and ftp client, you may immediately need to enter passive mode to succeed with ftp access. This is entered as follows:.
ftp> passive
Passive mode on
ftp>
)
- Move to the directory for the database to which you plan to upload (eg wdb)
ftp> ls
drwxr-xr-x 2 513 400 4096 Aug 19 2003 itb
drwxr-xr-x 31 513 400 4096 Jan 8 11:31 wdb
ftp> cd wdb
- Move to the directory for the pulse, (you can create this directory if it does not already exist).
ftp> mkdir 60933
ftp> ls
drwxr-xr-x 2 513 400 4096 Dec 8 10:58 60933
ftp> cd 60933
- Add the files
ftp> put jet_60933_0d.dat
ftp> put jet_60933_1d.dat
ftp> put jet_60933_2d.dat
ftp> put jet_60933_com.dat
2.-4. Checking Your Data Online and Making Your Data Live.
Before the data can be made live it needs
to be checked and scheduled for upload. To do this go to the password protected
http://tokamak-profiledb.ccfe.ac.uk/UPLOAD/.
Selecting the database to which you wish to upload discharges takes you to a table of the discharges which have already been uploaded to the transit area. Your uploaded discharges can be checked and the resulting reports can be viewed online from this webpage. When you are satisfied that your data satisfies these checks, you can schedule the discharges for upload to the live database. (NB There is a finite period of time during which it is also possible to uncheck discharges which you have mistakenly flagged for upload.)
2. Checking Discharges in the Transit Area
To run checks for any uploaded discharges:
(i) select checkboxes for the discharges to check on the upload webpage and then
(ii) click on the Generate Reports buttun, which runs the checking code
interactively and returns links to the reports, and a summary.
Please read these reports online and ensure that there
are no significant errors in your data before continuing. Feel free to email us if you do not understand any of the error messages.
3. Scheduling Upload from the Transit Area to the Live Database
Once the data has been checked it can be scheduled for upload, also from the
main upload page:
(i) select checkboxes for the discharges for upload
(ii) click on the Schedule Upload button.
(iii) please enter a useful comment for each discharge and your contact details. (These inform database users about new or modified data.)
(iv) it is possible to cancel a scheduled upload before the upload takes place. This is done from the main upload page by selecting discharges and clicking on the Remove Scheduled Upload button.
4. Upload to the Live Database
Discharges scheduled for upload will be uploaded automatically via a cron job, which runs at regular intevals to update the live data. (In extreme circumstances the administrators can run this job at any time.) Note that the upload procedure will fail to make your discharge live if the submitted data does not conform to the official variable lists and data formats. If the upload is successful the uploaded data and reports will be removed from the transit area. In the event of upload failure the data will stay in the transit area, and report files will indicate why the upload attempt failed.
Examples of Recent Submissions
See recent database submissions for examples on how to lay out file formats:
NB: you need access to the restricted area to get there!
Some Useful Contributed Software
Various Ufile read/write software:
- Stan Kaye's FORTRAN and IDL routines to write UFILES from TRANSP could be useful for data providers.
- Malcolm Walters FORTRAN routines to read and write more recent and more flexible 0D UFILES
(also useful for data providers.)
Various software to read/write MDSplus trees:
- Simple IDL Example reading a few database items into an IDL program
- IDL procedure to read all profile database data from a given discharge into IDL structures (originally from Martin Greenwald)
- Fortran Example reading a few database items into a Fortran program, with the makefile to compile with the Absoft f77 compiler (originally from David Mikkelsen)
- MATLAB Example reading a few database items into a MATLAB program (from Frederic Imbeaux)
- IDL example to write an MDSplus tree on the tokamak-profiledb.ccfe.ac.uk MDSplus server (Malcolm Walters)
- TCL script to generate profile database model tree files (Josh Stillerman):
- MDSplus read/write examples in f77, f90 hopefully coming soon ....

 This current version of the Profile Database Manual is under development. It is the definitive source of information on Profile Database data formats etc, and over the coming months we intend to incorporate information on reading and writing MDSplus trees to bring this documentation fully up to date.
This current version of the Profile Database Manual is under development. It is the definitive source of information on Profile Database data formats etc, and over the coming months we intend to incorporate information on reading and writing MDSplus trees to bring this documentation fully up to date.